Scientists discover new two-step technique for editing proteins
Researchers at the Rosalind Franklin Institute and Oxford University have discovered a new way of making changes to proteins – a group of biomolecules responsible for a vast array of important functions in the human body.
By using light as a catalyst, scientists were able to create a reaction with an amino acid called tryptophan that helps pave the way for editing the information inside a protein to control its activities. Tryptophan is obtained through our diets and is one of the building blocks of proteins.
Unexpectedly, the researchers also found that the chemical process involved in the reaction with tryptophan produces a common functional group in organic chemistry known as an aldehyde, or formyl group. This group is rarely found in proteins. Its creation has a number of useful consequences and applications, including the generation of a fluorescence that could allow scientists to track the onward progression of a living cell once the aldehyde is produced on its surface.
The new study is published in the journal ACS Central Science.
The technique builds on the team’s previous work exploring the use of light as a benign trigger for activating (or deactivating) biological function in proteins by making and breaking chemical bonds to alter the information inside. Medicine is the natural direction for much of this work, with conceivable applications in drug and vaccine development, as well as the therapeutic use of protein editing itself through the enhancement of ‘deficient’ proteins or the introduction of synthetic proteins into the body.
Professor Ben Davis, who leads the Next Generation Chemistry theme at the Franklin and co-authored the study, described the research as an exciting development. “One of the things we’re intent on exploring at the Franklin’, said Professor Davis, ‘is the idea of applying chemistry in living systems to help us observe and understand biological mechanisms. With this technique, we’re putting something very small and relatively benign – the aldehyde group – directly into a protein inside a cell to allow us to interrogate it from a functional point of view. This is very much what we’re about in the Next Generation Chemistry theme: a hopefully bold fusion of synthetic chemistry with biology.”
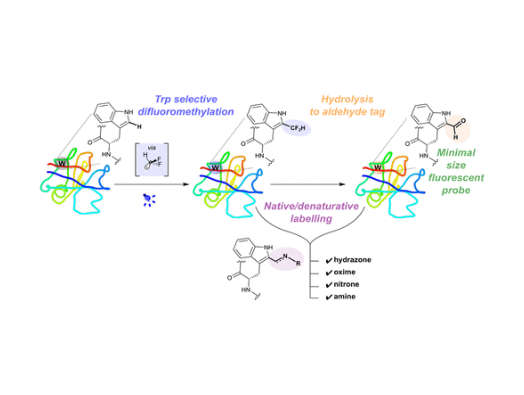
In this study, the researchers produced a particular type of reactive molecule known as a radical (specifically, a difluoromethyl radical), which under certain conditions selectively reacts only with tryptophan. This reaction can be used to make carbon-carbon bonds to the tryptophan that will be valuable in efforts to expand the range of outcomes achievable through protein editing.
By virtue of tryptophan’s ‘aromatic’ chemical environment (a description of its atomic structure named for the sweet odour produced by many similar compounds), the reaction with the difluoromethyl radical also becomes a neat (so-called ‘masked’) method of indirectly inserting an aldehyde group into a natural protein – the first time it has been done in this way.
Professor Davis added: “This two-step process unexpectedly led to a transformation that we have been trying to achieve for a number of years – so it’s a nice example of serendipity. The technique allows us subsequently to use conventional chemistry to attach a variety of molecules, or “labels”, to the aldehyde, enabling us to observe and track what the protein is doing, or to load it with “cargo”.
“It also creates a colour shift in the fluorescent spectrum of tryptophan. Because the change we’re introducing here is so small and benign, we can even label the surface of live cells – potentially one day in a more complex living system – and make them glow fluorescently. That allows us to track their movement in the same way we can with the traditional protein labelling, but instead here by manipulating endogenous fluorescence. This is a really exciting development and is right at the heart of our goals in gentle “editing”: applying small but effective chemical changes to give us a better understanding of biology.”
The research was led by Mateusz Imiołek, a junior scientist at the Franklin, and carried out in collaboration with Professor Véronique Gouverneur’s laboratory at the University of Oxford.
