Machine Vision for Bioimaging
Many biological questions can only be answered through visualisation. Seeing is believing, however seeing something that is biologically interesting usually also requires image processing to turn that qualitative observation into quantitative information.
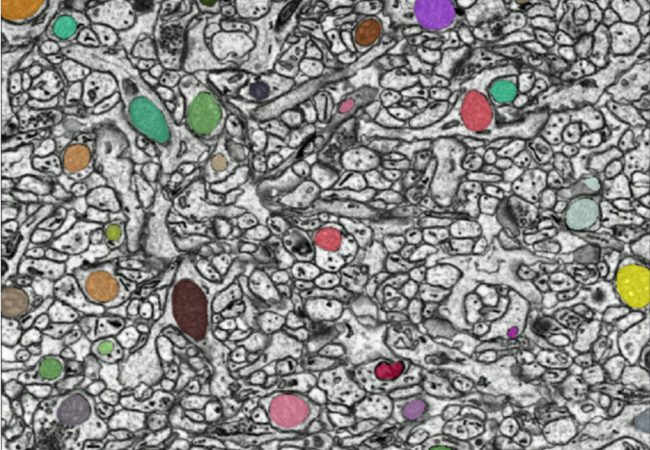
The first step in quantifying something about an image is to annotate it (annotation is the act of labelling an image or parts of an image). In order to speed up the science and automate image analysis, we need to generate and teach AI models to do this painstaking task but providing training data of complex 3D imaging data is manual and slow, there is very little of it currently available, so volunteers are sometimes asked to help.
Tools for efficient annotation of complex bioimage data
Annotation can be as simple as saying that there is a cell present in the image, or as complex as specifying the exact pixels that make up the mitochondria within the image. Annotating an image or a 3D volume can be very difficult and time-consuming.
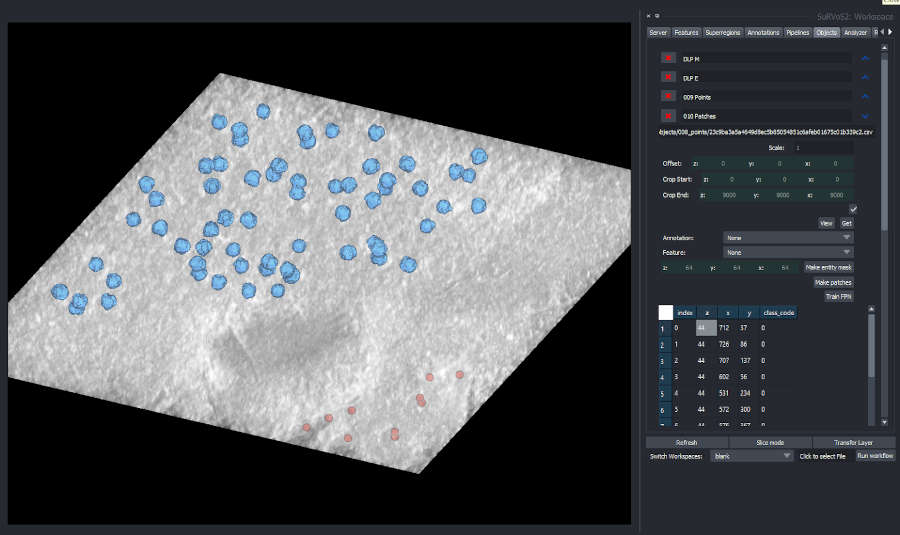
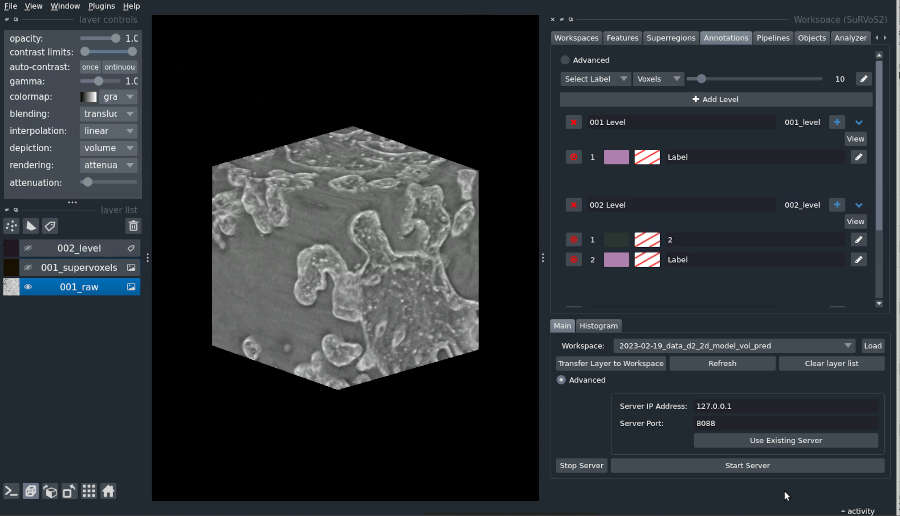
As an added complexity, the training of models for image analysis often requires an iterative approach and a lot of experimentation to steer the training process to produce a useful model. We develop tools to assist biologists in both the creation of annotation and the training of machine learning models. In particular, the open-source tool SuRVoS2 [LINK to Open Science] is designed for interactive, iterative workflows that allow experts to develop complex annotation and train deep learning segmentation models.
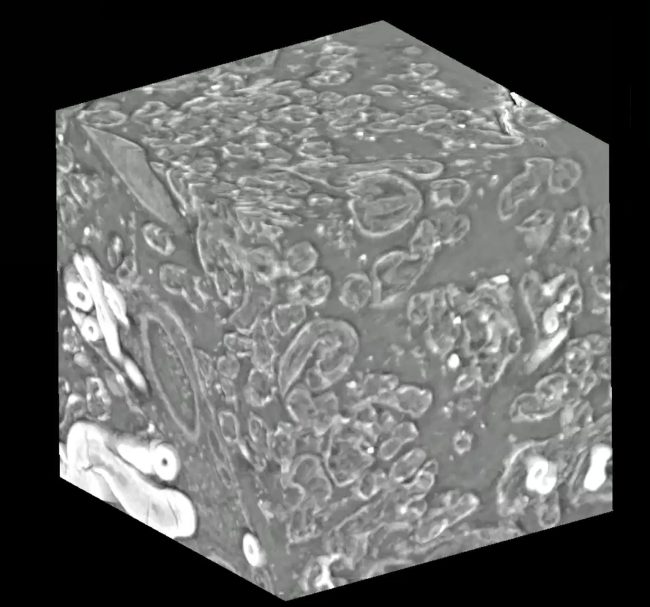
Using Deep Learning for Analysing New Imaging Modalities
Over the past decade, deep learning methods have become widespread in volumetric image analysis. Deep learning methods use Graphics Processing Units (GPUs) to train large neural network models with many layers. Many different neural network designs or architectures have been proposed, and different architectures have strengths and weaknesses for certain types of biological samples and for certain imaging modalities. Particularly important for research at the Franklin is the development of image analysis methods that can adapt to new approaches to microscopy.
Volume-segmantics is an automatic segmentation tool that is very effective for segmentation of biological volumetric data. To do this, it generates several predictions from a single raw dataset. This means that multiple outputs are provided, each with a slightly different prediction of which classes each voxel belongs to (e.g., inside of a cell vs outside of a cell). The last, essential stage is to combine all these different predictions to produce a final segmentation that best represents all of them together. One common approach to this is a maximum voting strategy where the class with the most “votes” wins. Though there are many alternative approaches one could take to combining predictions.
Instance Segmentation and Object-based Crowdsourced Annotation
Instance segmentation is a specialised image analysis approach we are researching at the Franklin that produces segmentations that detect objects in images. In many cases, it is not possible to analyse the images we take without time-intensive manual human analyses which would take too long for individual researchers to complete. In these cases, we turn to volunteers for help.
Tools to support object detection for volumetric image analysis are limited at present and we have extended SuRVoS2 to better support these tasks. Plugins that have been created to support object-based workflows include advanced connected component analysis with component filtering as well as mask rasterization and segmentation cleaning. Detected point sets can be visualised in the interface, and point-based annotation can be created and used to generate mask annotation for training instance segmentation models. In addition, we have developed crowdsourcing approaches to developing annotation for object-based analysis of bioimages and integrated tools for cleaning crowdsourced data into SuRVoS2. These various approaches and tools have been used in the analysis of viral infection and mitochondria distributions in human cells.