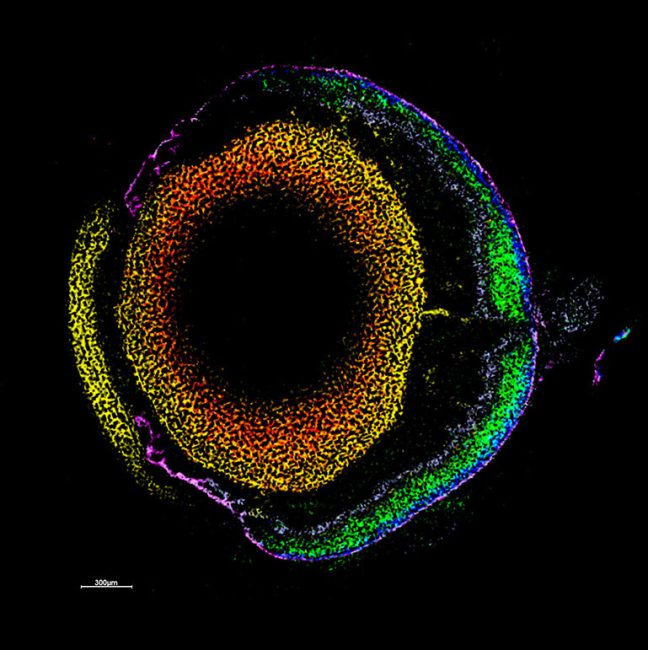
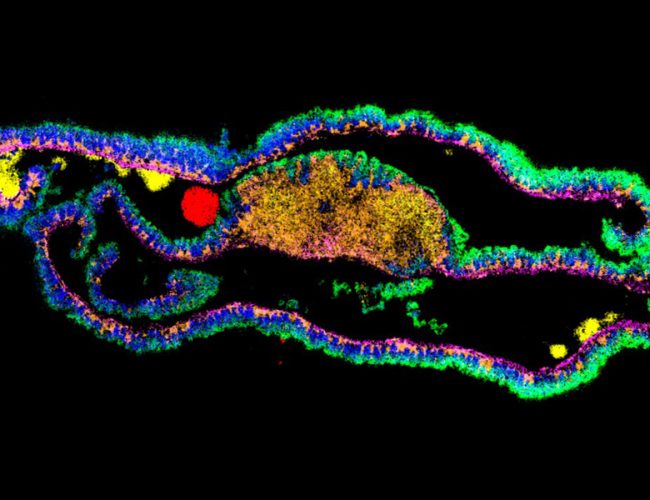
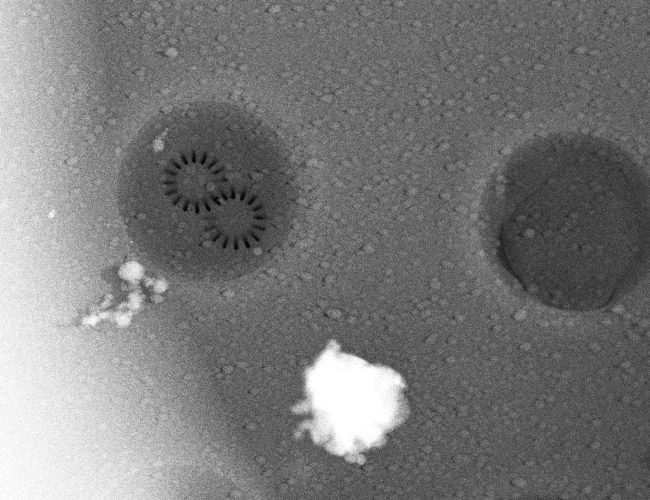
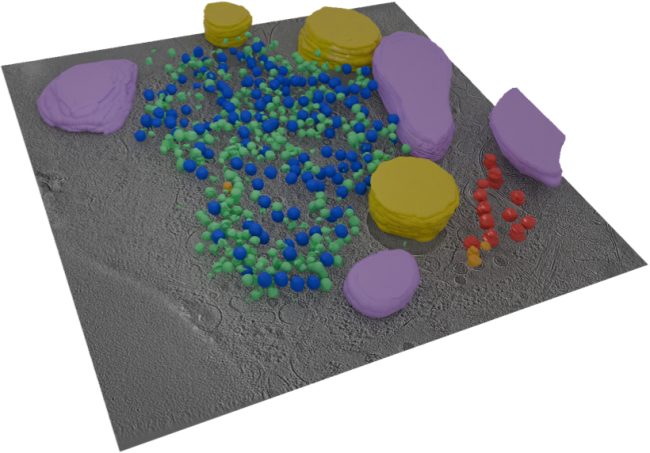
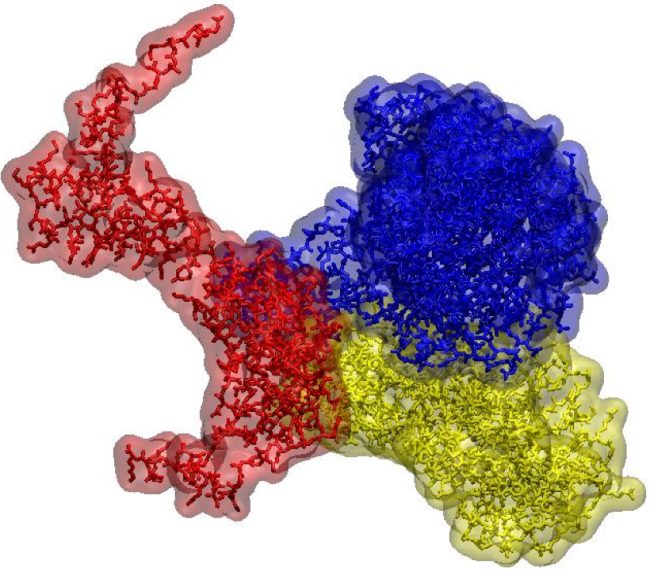
“The techniques we develop will have wide applications in the life sciences. For example, we want to create three-dimensional protein atlas of the mouse brain. Such a resource will enable new insights into neurodegeneration conditions such as Motor Neurone Disease.”
The ambition
These techniques give unique insights into the complete chemical biology of disease to improve future health.
In Motor Neurone Disease, for example, it is known that certain protein complexes accumulate in particular regions of the brain. Yet we don’t know enough about why these protein complexes accumulate in certain areas of the brain, or anything about the small molecule environment associated with them. By understanding these factors, we could get a much more comprehensive understanding of disease progression during neurodegeneration which could lead to new treatments in future.
Our work to make these techniques more robust, intuitive and holistic could lay the foundations for next generation equipment for molecular pathology.


What are we doing?
We are developing ways to unlock the most information possible from our biological samples. We plan to achieve this by bringing together mass spectrometry with other structural biology techniques, including high resolution microscopy, to map molecules across scales – from inside cells, to groups of cells, through to tissues and organs.
Mass spectrometry will give us valuable insights into the identity, characteristics, chemistry and environment of biological molecules, from large protein assemblies to small metabolites. We will also use computing and machine learning to speed up data acquisition and analysis to build three-dimensional maps of the molecules of life at different scales.
Why?
Traditionally our understanding of the spatial biochemistry of cells, tissues and organs has been limited to specific molecular classes. The techniques we are developing will give us highly detailed information about proteins, their structure and their broader molecular environment.
Research examples
We would like to create a three-dimensional protein atlas of the brain using mass spectrometry data. Alongside this, we will develop mass spectrometry approaches to understand the small molecule environment on a subcellular level. In the longer term, we would like to combine mass spectrometry imaging data, both small molecule and protein, with information from cryo-Electron Microscopy and Emission Tomography to get a more detailed understanding of cellular environment.
We aim to find new ways to share and visualise the data we generate with research teams across the UK and globally to improve our understanding of the biology of the brain, which could lead to new ways to improve health in future.
Why the Rosalind Franklin Institute?
The Institute is focused on building expertise to see life at different scales from molecular interactions, to cells, to tissues to whole organs. We have the capability to effectively zoom in and out to see life at different scales, as well as to layer multiple imaging techniques to give broader information to better understand biology.
Researchers at the Institute have access to state of the art mass spectrometry and innovative sample preparation techniques. Engineering and computing expertise will ensure that the techniques we develop become more robust and automated, which will speed up our research and make the techniques more likely to be adopted by others. This challenge will generate large amounts of data which will benefit from the Institute’s expertise in data science and AI.