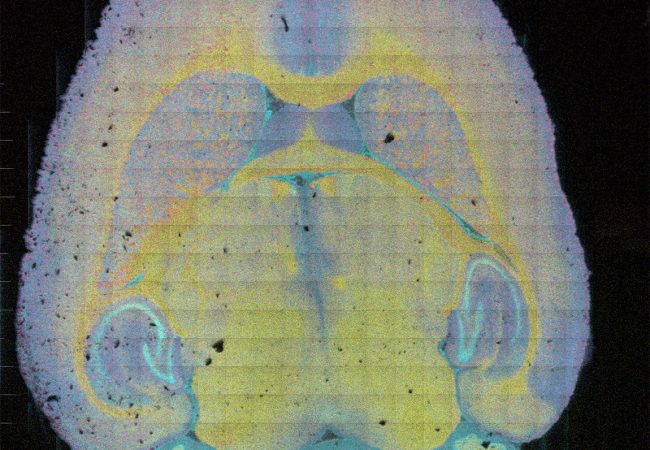
This would represent a step change in capability and will require technological advances in every part of the instrument – from the ion beam to the optimisation of post ionisation and next generation detectors, as well development of a multi-omic workflow to enable a complete chemical picture for a given biological system.
Subcellular Imaging
Next generation MS instrumentation will enable rapid molecular mapping of cells in tissue enabling elucidation of the chemistry behind biological mechanisms.
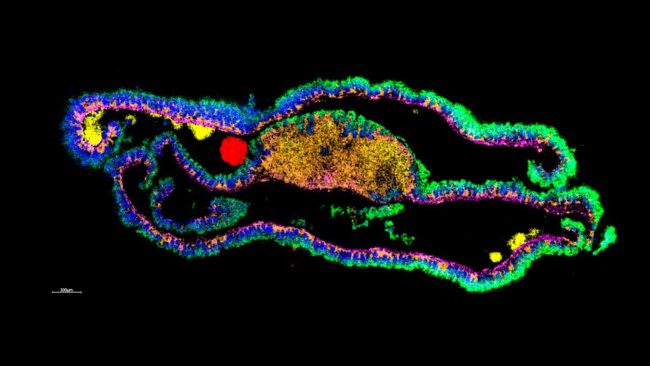
To improve spatial resolution in mass spectrometry imaging (MSI) will require technological breakthroughs in sensitivity with optimisation of post ionisation, new ion beams and laser modalities and improved ion optics and transfer. Additionally, for MSI to become routine, high speed, multi-scale, in situ, and targeted additional instrumentation and techniques are necessary. In collaborations across the Franklin, we will also make improvements in the types of samples we can analyse, through: nanobody tagging; cryo-analysis; cross comparison and integration with SEM; dynamic (time resolved) SIMS and DESI. These advances for MSI need to made in tandem to intensify the rate of progress:
- Next generation detectors for microscope mode MSI. Stigmatic ion microscope imaging enables us to decouple the primary ion (PI) beam focus from spatial resolution and is a promising route to attaining higher throughput for mass spectrometry imaging (MSI). By coupling the beam with a position-sensitive spatial detector we can achieve MSI through simultaneous desorption of ions across a large field of view, enabling images to be recorded over an area of 5 mm2 in a matter of seconds. Presently using time resolved optics we can achieve <5 µm spatial resolution with >5,000 mass resolution. Further development will make it possible to improve this performance for even better sensitivity, speed and resolution.
- Development and optimisation of post ionisation to enhance sensitivity to enhance sub-cellular spatial resolution
- Exploration of novel desorption and ionisation sources to give optimal sensitivity and spatial resolution with minimal fragmentation including: fs and ns IR lasers; development of water ion beams (single droplet/pulsed/multiply charged).
The complexity of human biochemistry requires multi-omic workflows to enable a complete chemical picture for a given biological system. We aim to enable a holistic multimolecular approach to resolve the biochemical complexity of human pathology by uniting previously disparate techniques and sample preparations together to provide a truly world-class suite of multi-omics tools.
Spatial analysis of isomeric compounds with TIMS
We are developing methodology to analyse the vital and highly challenging glycan subclass: glycosaminoglycans (GAGs). By developing tissue preparation methods to enable molecular release and selectivity, optimisation of ionisation source for soft(er) ionisation and the separation and identification of isomers with TIMS, we are building a new toolkit for deep, spatially resolved glycomics analyses.
Exploration of multiple fragmentation modes
When analysing of a broad range of molecular types, standard fragmentation methods are often sub-optimal. Through use of photonic and electron-based fragmentation, coupled with gas phase TIMS separation, we are developing tools to characterise difficult compounds on our prototype TIMS-FTICR instrument and then identify them in tissue.
Unifying different spatially resolved ‘omics
When analysing a particular molecular sub class, multiple considerations must be taken into account. These considerations often affect the ability to analyse other sub classes in tandem, yet to garner the maximum chemical information from the tissue, analysis of multiple molecular sub types is required. Hence, we are investigating ways to unite disparate methodologies, through the development of new sample preparation modes, the use of novel ion sources such as REIMS and with modifications to old ion sources.
In situ fragmentation analysis for macromolecular identification
Traditionally, fragmentation for structural elucidation in MS has been carried out using CID or chemoenzymatic digestion. Both of these are limited and struggle when being used in an imaging modality (due to speed or molecular delocalisation). We are seeking new modalities that enable on-tissue fragmentation without delocalisation such as solid-phase digestion of analytes with plasma or mixing SIMS modalities to explore inherent fragmentation.
Ionoptika Ltd
The instrument developed in collaboration with UK based Ionoptika enables a dramatic improvement in mass spectrometry imaging (MSI). The new instrument is capable of imaging the whole surface simultaneously, allowing more accurate imaging at faster speeds than was previously possible.