MS, however, currently does not provide enough information that would allow full structural elucidation, and our project develops a computational and experimental toolkit to overcome this limitation.
Mass Spectrometry (MS) is an extremely sensitive analytical technique, but full structural characterization of small molecules (ranging from metabolites to lipids and peptides) using MS data only remains a huge challenge and a major bottleneck in a number of areas like metabolomics, glycomics and natural product chemistry.
To address this issue, we are developing computational and experimental strategies to assign molecular structures to spectra based on 3D structural interpretation of MS, tandem MS, and Ion Mobility Spectrometry (IMS) data. We compute accurate gas-phase structures, predict fragmentation chemistries and develop strategies to harmonise MS with other analytical techniques like microED, NMR and gas-phase ‘action’ IR spectroscopy.
To predict fragmentation chemistry, we use our ‘Universal Fragmentation Model’ (UFM) that combines gas-phase ion chemistry and modelling to generate and test dissociation hypotheses. A unique feature of the UFM is that it can predict rearrangement chemistries and therefore validate complex fragmentation behavior. The UFM is harmonized with our computational strategy that can rapidly determine accurate gas-phase structures to facilitate interpretation of IMS and IMS-MS/MS data. Our work also aims at developing new ML-based structure elucidation techniques that are built on chemically interpreted MS data.
As a technology platform we intend to reduce the current MS vs. NMR information gap while keeping the sensitivity of the former. Our technologies are used to discover new disease biomarkers and characterise natural products and deeper understand biochemistry. Further applications aim at supporting drug development strategies.
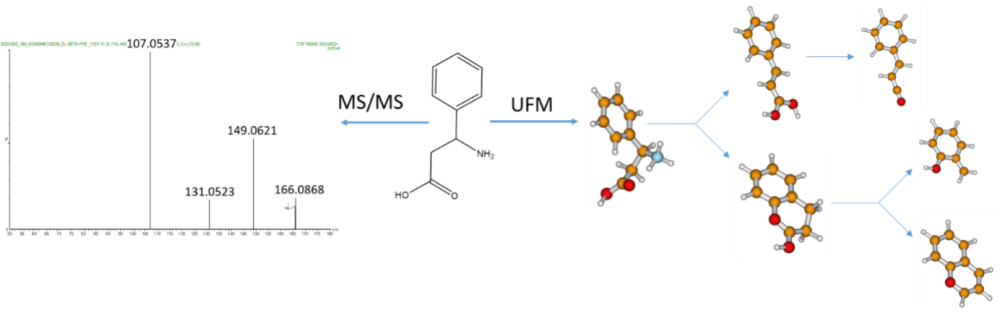
Our team is keen on collaboration making use of our unique computational capabilities for any MS-based projects in the UK and worldwide.





