The Franklin provides expertise in segmentation and quantification of three-dimensional X-ray images of the placental tissue and vascular network.
In Utero
Stillbirth is the result of varied processes taking place within both the mother’s and baby’s organs during pregnancy – a time of rapid changes.
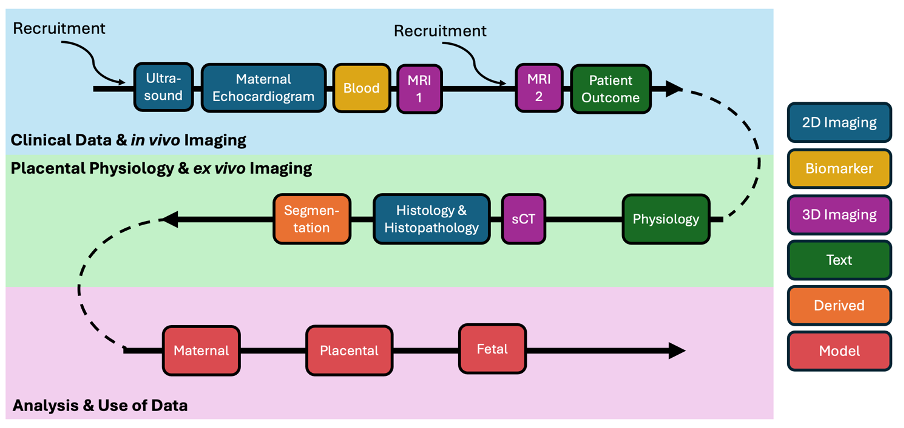
This project links medical and research outcomes across imaging approaches, time and patients, integrating this data within a computational model to simulate blood flow within and between the mother, placenta and baby (Figure 1): the ambition is to provide tools for predicting pregnancy outcomes and testing possible interventions.
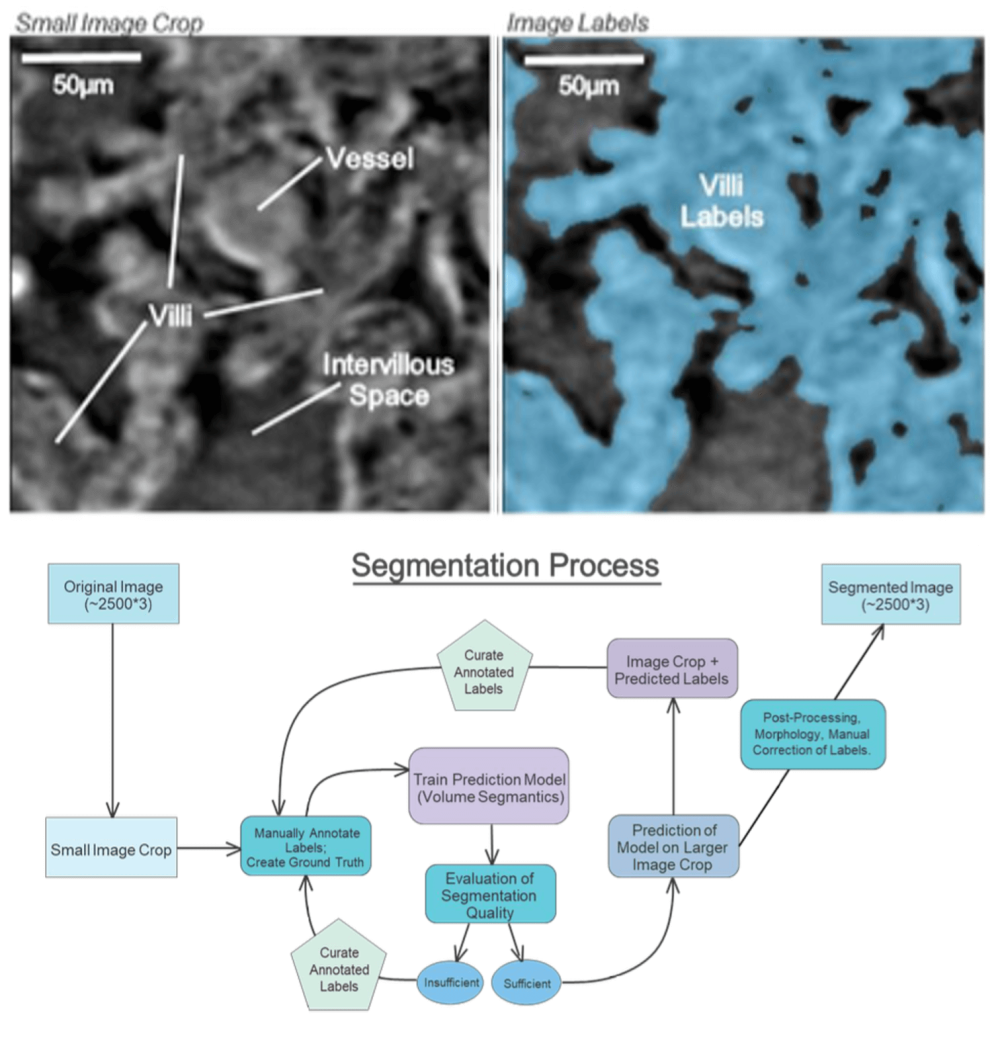
To understand and quantify 3D placental villous structure so it can be related to organ and system level function, synchrotron CT is undertaken (Pešić et al. 2013; Rau et al. 2011; Tun et al. 2021). A robot autosampler is employed for autonomous sample changes allowing up to 112 samples to be loaded per session. Generally, three scans are taken of each pole of placental tissue for a total of 12 or 16 scans per placenta (single- or dual-sided perfusion). Each scan is approximately 50 GB and represents a field of view of approximately 2 mm x 2 mm x 1.8 mm (Figure 2 top left).
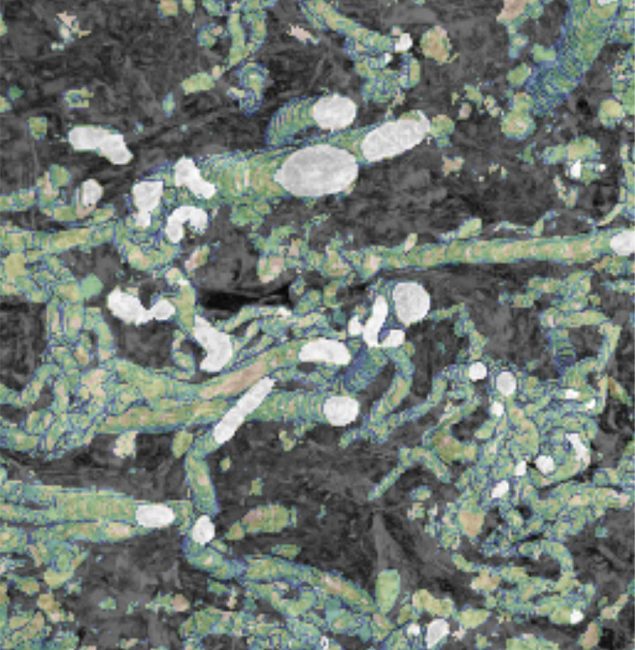
The anatomical complexity of sCT datasets makes automated segmentation critical to enable quantification of meaningful numbers of patient samples. To achieve this, scans are evaluated for quality including size and severity of artefacts. High-quality scans are segmented using a U-Net Python-based framework, implemented as an extension of the PyTorch Volume Segmantics package (Bellos and Basham 2022; Figure 2 top right). Annotator time is minimized using a pseudo-labelling approach whereby a model is trained on a dataset of manually annotated samples and then is used to predict a region of interest, ROI, from a new scan. This prediction is manually corrected and then used to train a new model. In addition, the segmentation of some scans is predicted using a model trained using a collection of ROIs from different scans. Napari (Sofroniew et al. 2024) and the SuRVoS2 Napari plugin are used for manual annotation (Pennington et al. 2022). Segmentation quality on a set of initial samples was above 0.90 dice score compared to manually annotated expert annotation. In a small number of segmentations some manual correction is needed. Outputs from sCT segmentations include measurements of maternal intervillous space, IVS, (ball and stick representations, area fraction), fetal tissue (area fraction), and vascular networks (skeletonisation, diameter, branch numbers, tortuosity).