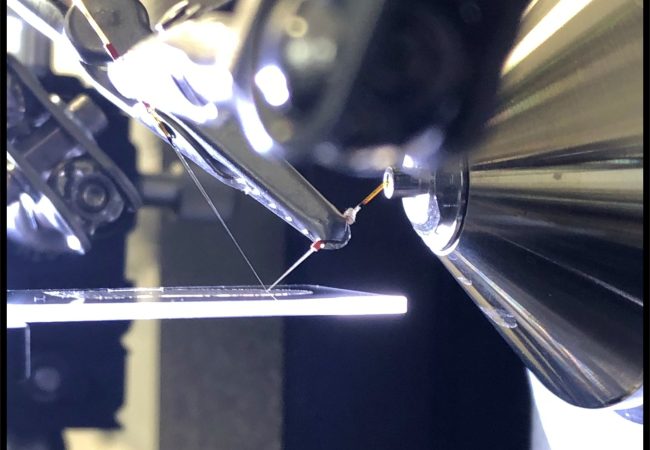
Its unique feature is the ability to probe protein interactions directly from tissue in a label-free and spatially-resolved manner.
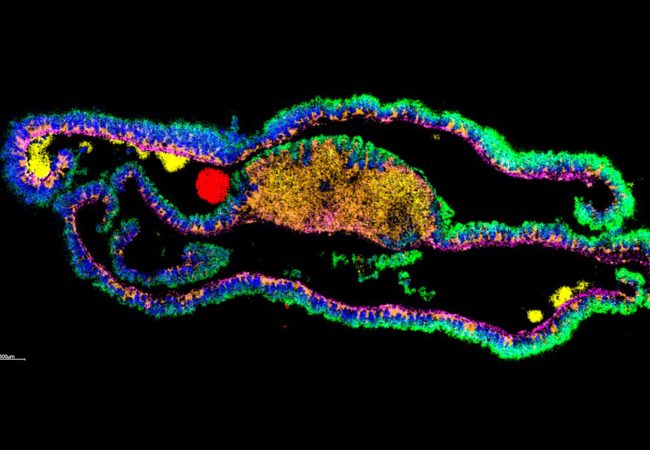
3D Protein Atlas of Brain
Native ambient mass spectrometry (NAMS) is an emerging technology which offers unprecedented potential for integration of spatial and structural biology – it promises major advances in molecular pathology and drug discovery.

Our aim in this project is to combine native ambient MSI with data science to generate a 3D protein brain atlas as a molecular pathology resource for the broader scientific community. Our research will focus on developing the hardware and software tools required to deliver this ambitious goal. We will work to make NAMS a highly-sensitive, robust and automated tool, capable of generating large numbers of serial 2D imaging datasets that will form the basis of the 3D atlas. In parallel, we will develop data analysis and visualisation tools to construct and interrogate the atlas.
NAMS combines several branches of mass spectrometry: native mass spectrometry, in which non-covalent interactions present in solution phase are maintained into the gas-phase i.e. proteins remain folded and complexes remain intact; ambient mass spectrometry, in which thin tissue sections are sampled directly under atmospheric conditions following no (or little) sample pre-treatment; mass spectrometry imaging, in which the spatial distribution of analytes is visualized; and top-down mass spectrometry, in which the identity of a protein can be deduced from its sequence fragments. NAMS is capable of identifying and imaging protein assemblies, including membrane protein assemblies, endogenous protein-ligand complexes and metal-bound proteins, and protein-drug complexes formed in vivo.
NAMS involves different modes of tissue sampling to support different goals. In this project, we are focusing on nanospray desorption electrospray ionisation (nano-DESI) for native ambient mass spectrometry imaging (MSI). Native ambient MSI is a label-free molecular imaging technique with the unique capability to identify and map the distribution of endogenous protein complexes within tissue sections. Fresh-frozen tissue is analysed intact, i.e., there is no requirement for homogenization, nor is there a requirement for the development of specific antibodies or tags.
Thermo Fisher Scientific
Researchers at the Franklin have been working closely with Thermo Fisher Scientific through our Wellcome funded ‘Electrifying Life Sciences’ programme to develop the next generation of electron microscopes for imaging the smallest structures of cells in intact tissues, bring these tools to market and establish new methods and standards for the community to enable their wide use.