“We will develop game changing techniques to map out the biology of our organs. We will begin by studying the placenta to understand what causes it not to function effectively in some circumstances. Our hope is to use computational modelling to understand better how to reduce stillbirth by identifying women most at risk and improving treatments to reduce babies’ exposure to hypoxia.”
The ambition
The techniques we develop will help answer tricky medical questions, such as how to predict which women are at highest risk of stillbirth. We hope that our computer models will help to understand how drugs would affect the mother-placenta-baby system to influence clinical practice. To future proof our new technologies for use in the clinic, we will work at an early stage with regulatory agencies and industrial co-development partners.
We also hope the imaging and image analysis techniques we pioneer will be adopted by the research community more broadly and applied to other health challenges to better understand human health across scales.


What are we doing?
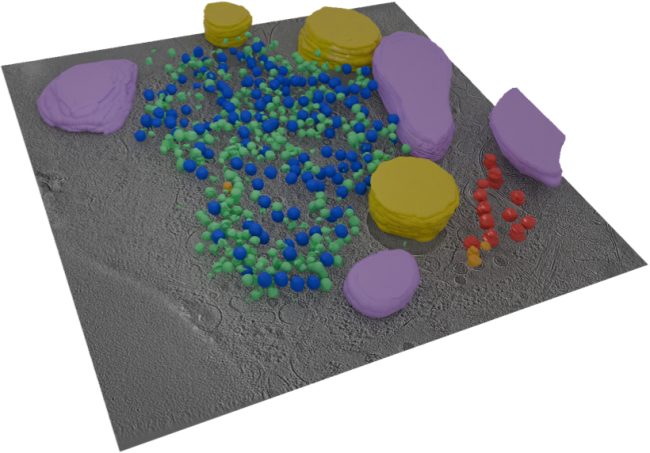
We are bringing together many imaging techniques including MRI, synchrotron CT, X-ray microscopy, electron microscopy, and mass spectrometry, with computing and machine learning, to map features of cells, tissues and organs in the same sample. This information will be used to create simulated systems to better understand disease.
Why
Current clinical imaging approaches have low resolutions (cm to mm) but large fields of view. Diagnostic pathology and pharmacological applications demand high resolution (mm to multiple Ångstrom) data. This and major differences in sample preparation limit the routine integration of these different classes of information. Developing ways to bring together a variety of three-dimensional techniques will allow us to see cells and tissues in greater detail combining information across scales in simulated systems, including where they sit in the wider context of the tissue or organ. Overcoming these barriers could transform clinical diagnostics and early stages of new therapeutics development by providing vital information about mechanisms of disease and informing the development of new treatments or improvement of existing treatments.
Research Examples
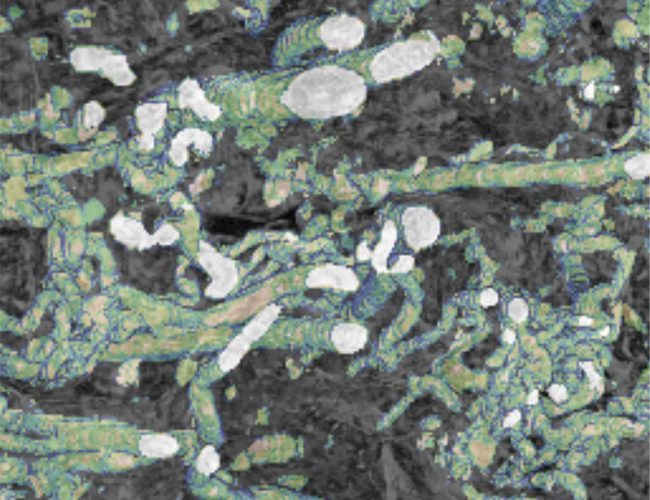
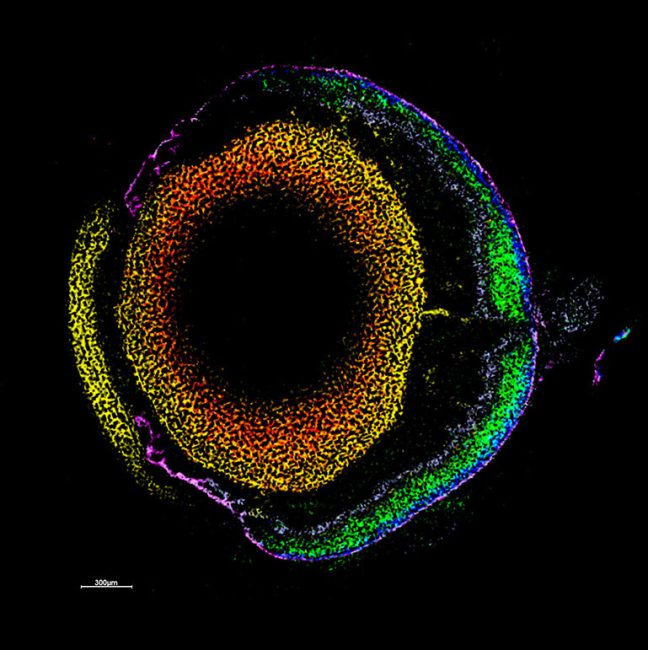
One of the flagship projects for this challenge is imaging human placenta to better understand how placental health determines outcomes in birth. We are working with a coalition led by the University of Manchester Research Hospital, studying the placenta during and after pregnancy. After birth, we examine the placenta using innovative imaging techniques, such as synchrotron CT at Diamond Light Source, to produce three-dimensional maps of tissue and blood vessels. We then use the data generated to build computer models that simulate how the mother, placenta, and baby interact and may be affected in different situations which could potentially lead to better prediction of pre-eclampsia, fetal growth restriction or stillbirth. We hope our findings will help to understand what abnormalities in the placenta might contribute to harmfully low levels of oxygen for the baby.
In the longer term, the techniques and approaches we develop could also be used in other medical contexts, for example, to help understand endometriosis and the long-term impact of infection on health.
Why the Rosalind Franklin Institute?
This research is pioneering and requires a highly collaborative multidisciplinary approach that includes clinicians, researchers experienced with different leading-edge imaging techniques, and computer scientists working closely together. This is research that can happen more easily and quickly because of the Franklin’s location on Harwell Campus and specialised expertise and equipment.