Exploration of unwanted immune response to penicillin lays groundwork for better drug design
Around 6% of people in the UK are allergic to penicillin according to their medical records – the most commonly reported drug allergy.
And although research suggests the majority of those patients could, in practice, tolerate the drug, penicillin allergy at all levels has a big impact: those with “true” allergy may be at risk of death through anaphylaxis, while diagnosis of penicillin allergy is associated with higher total numbers of antibiotic prescriptions, undermining antibiotic stewardship goals and increasing the risk of antibiotic resistance.
Yet, surprisingly, we do not yet know the full details of the biological mechanisms underpinning the body’s reactions to these small molecule drugs – why our immune systems decide to produce antibodies against a helpful antibiotic, potentially sparking an allergic reaction or possibly reducing the drug’s effectiveness therefore remains a key question.
New research by scientists at the Rosalind Franklin Institute and the University of Oxford, funded by the Rosetrees Trust, digs into these mechanisms, providing the beginning of a roadmap for predicting unwanted immune responses or even developing antibiotics and other small molecule drugs that could avoid them altogether.
The study’s lead author is Dr Lachlan Deimel, now of the Rockefeller University in New York, who worked on the research while undertaking his PhD with Professor Ben Davis at the Franklin and Professor Quentin Sattentau of Oxford University’s Dunn School of Pathology. Dr Deimel says: “The immune system has to make decisions about what foreign bodies it raises antibodies against. We have a good idea in broad terms of what is likely to provoke an antibody response, but we don’t have a deep understanding of the molecular rules that govern those responses.”
Penicillin is part of a group of antibiotics called β-lactams, which refers to a moiety in their chemical structure. This moiety has a reactive property that causes the drug to covalently bind not only to the enzyme that is its antibiotic target but also potentially indiscriminately to proteins within the body, in turn provoking the immune system to respond – such as producing antibodies usually associated with fighting off disease.
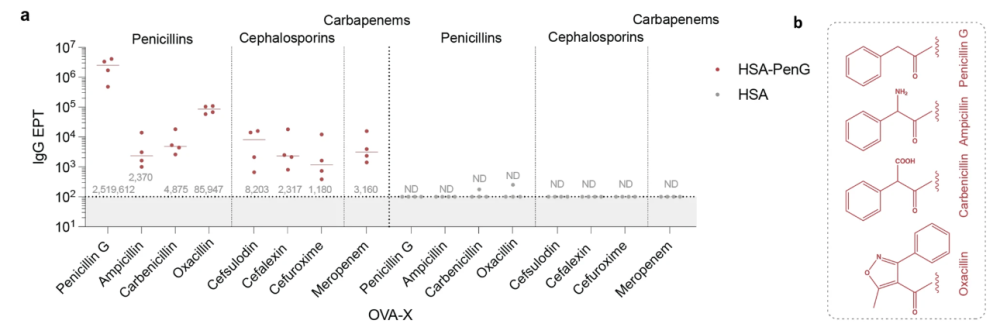
The interdisciplinary team of researchers set out to unpick the processes and trends behind this antibody response. Using mice as a model organism, they explored the “pharmacokinetic” conditions necessary to elicit an unwelcome immune reaction. Length of exposure to the drug was identified as one key aspect – itself influenced by factors including drug clearance rate, dosage and route of administration – while another was the base level of immune activity at the time of exposure.
More conceptually, the researchers then tackled the question of whether the antibodies produced could be predicted through a molecular rationale – and therefore whether, in theory, drugs could be engineered to avoid binding to these predictable antibodies and thus become less immunogenic.
Dr Deimel says: “There are clear translational implications of this work related to drug design, from the allergy aspect to the pharmacokinetic elements, but at its heart this is about exploring the deep, basic science – something the Franklin does really well. How does the chemistry of a small molecule drug affect its propensity to fasten to proteins in the body and drive a downstream reaction of antibody production? How do we stop that from happening? Immunology is fundamentally a chemical process, with products like antibodies reliant on things like their molecular structure and the biophysics of their interactions with other agents to be able to do their job.
“We’ve been able to increase our understanding of the molecular underpinning of certain antibodies in mice, which lays the conceptual groundwork for thinking about reverse engineering non-allergenic drugs. It’s not a perfect template because it’s in mice, but it’s a template nonetheless – that’s the most exciting part for me.”
Professor Quentin Sattentau of Oxford University, one of the paper’s senior authors, adds: “It is the power of our interdisciplinary approach, combining chemistry with immunology, that has allowed us to address this important question.”
Professor Ben Davis, as the Science Director of Chemistry at the Franklin, sees this investigation as part of a broader ability to explore the physiological consequences of such in vivo chemistry: “Our ability now to characterize the reactive properties of covalent drugs in living organisms not only allows us to unpick the direct pathological consequences, as we saw here, but also paves the way for the diverse use of covalent bond-forming and bond-breaking chemistries in a precise way to probe and understand biological function in context.”
