This year we are celebrating 5 years since the Rosalind Franklin Institute became a legal entity. The Franklin was established to develop the next generation of technologies for life science, to help solve the grand challenges in human health and disease.
In the past five years, we have made great head way in achieving this aim – we’ve turned a business plan into a building, which was delivered on time and on budget, and created a team producing world class science. We have also set-up a unique PhD programme with excellent students completing highly interdisciplinary and collaborative projects.
Our science teams have made excellent progress in the first stage of the Franklin, below are just a few highlights from the past 5 years.
1. Electrifying Life Sciences
The Electrifying Life Sciences project is a flagship project for the Franklin, which aims to develop pseudo-atomic level imaging technologies for cells and tissues. The project is run in collaboration with Thermo Fisher Scientific and is funded by Wellcome.
Our multidisciplinary team of scientists have shown, for the first time, that the imaging technologies developed can produce high-quality volumetric information of natively preserved biological samples, precluding the need for fixation or resin embedding. This paves the way for mesoscopic maps to be produced of whole cells and tissues in pristine detail.
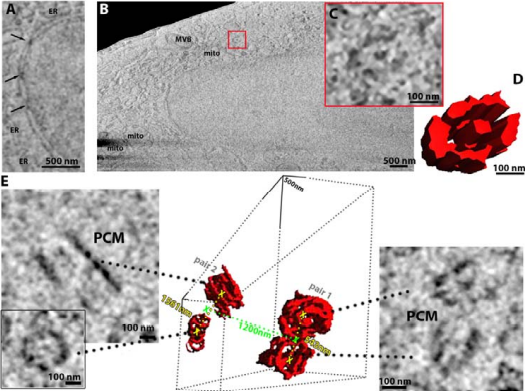
Serial pFIB/SEM volume acquired of a HeLa cell, using argon for 1115 milling imaged 52° to the surface by SEM.
Using frozen-hydrated native samples allows the maintenance of ultrastructural detail, and by employing machine learning and informatics-based tools to help refine the technique, the team has developed an imaging method which, in combination with other technologies at the Franklin, could be used by clinicians to analyse both healthy and diseased tissue at the molecular level. This work was published in eLife – read more about it here.

Ribosome structure
The team used a new instrument, a prototype version of the recently announced “Arctis” microscope, to obtain the structure of a human ribosome to 4.9Å – achieving the highest resolution structure within a mammalian cell. This achievement reflects the improved preparation process and capabilities of the microscope. This work was published in Nature Communications – read more about it here.
2. Zero sized labelling
Researchers at the Rosalind Franklin Institute and Oxford University have discovered a new way of making changes to proteins – a group of biomolecules responsible for a vast array of important functions in the human body.
By using light as a catalyst, scientists were able to create a reaction with an amino acid called tryptophan that helps pave the way for editing the information inside a protein to control its activities. Tryptophan is obtained through our diets and is one of the building blocks of proteins.
The researchers also found that the chemical process involved in the reaction with tryptophan produces a common functional group in organic chemistry known as an aldehyde, or formyl group. Its creation has a number of useful consequences and applications, including the generation of a fluorescence that could allow scientists to track the onward progression of a living cell once the aldehyde is produced on its surface.

Tryptophan editing through the production of an aldehyde tag.
Professor Ben Davis, Science Director for Next Generation Chemistry and co-author of the study, said, “Because the change we’re introducing here is so small and benign, we can even label the surface of live cells – potentially one day in a more complex living system – and make them glow fluorescently. That allows us to track their movement in the same way we can with the traditional protein labelling, but instead here by manipulating endogenous fluorescence. This is a really exciting development and is right at the heart of our goals in gentle “editing”: applying small but effective chemical changes to give us a better understanding of biology.”
The study is published in the journal ACS Central Science – read more about this research here.
3. World first time-domain electron microscope for life science
The Franklin is home to a ground-breaking electron microscope. The microscope was manufactured by JEOL ltd, and the Franklin team is working closely with the JEOL team to refine the machine so it can be used for imaging biological materials.
The microscope, known as ‘Ruska’, will work with biological samples both cryogenically frozen and in liquids, which will enable imaging of molecules in motion. By combining high speeds with liquid cells, the Ruska microscope will be able to ‘film’ proteins as they fold or image drugs interacting with other molecules. For cryogenically frozen samples, the frames captured as the beam passes through the sample will enable the creation of 3D models of biological structures, such as viruses or proteins.
4. New and unique mass spectrometry instrumentation
The Biological Mass Spectrometry team have been working in partnership with Bruker, Waters and Ionoptika to develop the next generation of mass spectrometry technologies for the analysis of biological samples. The group’s ultimate goal is being able to monitor and analyse what is happening at the molecular level in every type of cell at every time – called functional proteomics.
The Bruker instrument is designed to be a precision tool for the structural study of biomolecules. In biology, shape and function are closely linked, so studying the shape of biomolecules tells us how they might work, and how we can bind drugs to them if they’re linked to a disease.

The installed TIMS-MRMS
This new TIMS MRMS instrument combines trapped ion mobility spectrometry (TIMS) with the magnetic resonance mass spectrometry (MRMS) technique. The TIMS cell probes the shape of biomolecules, while the MRMS lets us fragment biomolecules in many different ways to study and elucidate aspects of their internal structure. The MRMS detector based on Fourier transform-ion cyclotron resonance (FT-ICR) technology has unparalleled mass resolution and accuracy, so we can differentiate between very similar mass biomolecules that other instruments wouldn’t be able to resolve.
Find out more about this instrument here.

The prototype stigmatic imaging secondary ion mass spectrometry (SIMS) instrument
The stigmatic imaging SIMS instrument developed in collaboration with Ionoptika will enable rapid molecular mapping of biological tissues at unprecedented speed, as this type of mass spectrometry imaging decouples acquisition time from spatial resolution. Typically, mass spectrometry imaging scans across a surface taking a mass spectrum at each spot to build up the pixels of the image. In this case, however, the whole surface is imaged simultaneously using state of the art cameras that operate as an array of position and time sensitive detectors that record a mass spectrum for each pixel in the camera image. It therefore represents a promising route to attaining higher throughput.
5. Building Scientific Computing Infrastructure and Key Collaborations
The Franklin’s Artificial Intelligence and Informatics theme has developed the key scientific computing infrastructure at the Franklin and set-up crucial collaborations to allow scientists to maximise their data.

Baskerville
One such collaboration is a project called Baskerville, which is run by the University of Birmingham in collaboration with The Rosalind Franklin Institute, The Alan Turing Institute and Diamond Light Source. This project has developed a computing system aimed at helping researchers speed up the scientific discovery process and provide new insights into important research questions.
Find out more about Baskerville here.
Another key collaboration is with the general public – the AI theme have been harnessing the power of the crowd to help them analyse the huge quantities of data collected by the Franklin’s state-of-the-art imaging. The AI team use the crowd sourced data to train their machine learning algorithms which will saves scientists a lot of time, as this data would take months or even years to analyse.
The next five years
During the first five years of the Franklin, we have built our technologies and we are now turning to the next phase of the Franklin where we will continue to push and apply these technologies to solve real biological problems. The Franklin will be dedicated to bringing about a revolution in imaging which will allow us to see entire cells and tissues to pseudo-atomic level detail.
Viewing biomolecules in their native context of the cell will be enormously beneficial for guiding the discovery of new medicines and diagnostics. We have already seen the benefit of studying individual biomolecules in improving health, well-being, and prosperity. This reflects the unique power of atomic level mechanistic insight to guide the discovery of new medicines.
However, the isolated molecule approach when used against complex multifactorial diseases such as neurodegeneration, AMR infection, aging and chronic inflammatory conditions that afflict our society has been less successful. The era of ‘easy’ targets is at an end.
We can only find the solution to these problems by understanding diseases at the atomic level within the relevant cells and tissues – “atomic pathology and physiology”. This approach leverages the established power of atomic mechanistic insights, but by making them in the biological context we can capture information and interactions which can only be seen in the organism, and are key to truly understanding function.
The technology needed to deliver the era of atomic pathology and physiology does not exist; the Franklin is tasked to develop and apply them. Success will lead to a new wave of economic, societal and health benefits to the UK.