A multidisciplinary team of scientists has shown, for the first time, that a microscopy technique called cryo-plasma FIB/SEM can produce high-quality volumetric information of natively preserved biological samples, precluding the need for fixation or resin embedding.
This paves the way for mesoscopic maps to be produced of whole cells and tissues in pristine detail.
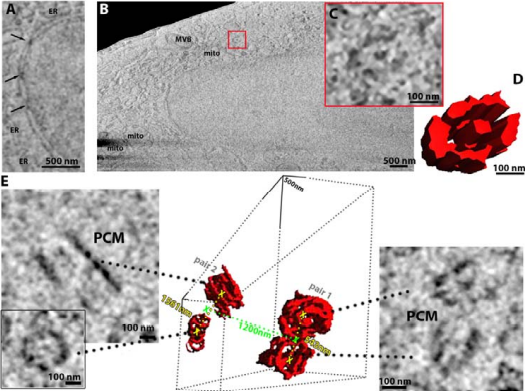
Using frozen-hydrated native samples for maintaining ultrastructural detail, and by employing machine learning and informatics-based tools to help refine the technique, the team has developed an imaging method which, in combination with other technologies at the Franklin, could be used by clinicians to analyse both healthy and diseased tissue at the molecular level.
The researchers hope their work – which has achieved resolution of biological material at the nanometre scale – will lead to new insights into the molecular causes of conditions such as Alzheimer’s disease, heart disease, kidney malfunction and psoriasis.
The study is published in the journal eLife.
Dr Michael Grange, Group Leader at the Franklin and one of the paper’s senior authors, said: ‘The idea of this paper was to explore whether we could keep biological samples in a frozen, hydrated state while still being able to produce large volumetric information with high detail. Such a method allows us to further probe how we can feed this into higher-resolution analysis using cryo-electron microscopy. The context of our study with regard to potential benefits of using plasma beams of noble gases rather than the traditional gallium, such as greater milling rates or reduction in implantation, also means that we were able to demonstrate their general applicability for in-situ structural and volumetric approaches. In theory, this dual approach would allow us to determine the structure of very small biological features with a high resolution. This represents a step-change in the way we perform structural biology.’
The team found the technique to be successful across a range of samples – bacterial organisms to human cells, as well as mouse heart and brain tissue – using a range of plasma gases, characterised in the paper.
Dr Grange added: ‘Although cryo-plasma FIB/SEM isn’t a new technique, what makes this exciting is that we’ve used cryogenic samples, unperturbed in their native state, and achieved resolution in the tens of nanometres. We hope this technique can be adopted in the clinic to allow analysis of human tissue at a smaller scale than previously available, helping to determine what’s happening at the molecular level in health and disease by comparing both types of tissue. We also want to drill down through the resolution scales even further – going from nanometre to sub-nanometre.’
Dr Maud Dumoux, Technology Lead for Cryo-Imaging at the Franklin and the paper’s lead author, said: ‘The Franklin is keen to make the most of the time a sample spends in a microscope. As milling is used to thin the sample to the desired thickness for subsequent imaging, such as cryo-electron tomography, linking it to high-resolution SEM imaging helps our understanding of the sample by putting it back into its context. By linking the different scales, we bring together different disciplines across life science – in this case cell biology and structural biology.
‘At the Franklin we have the opportunity to access the latest technologies, and we aim to make them accessible to wider communities. This paper presents a new plasma source utilising a range of gas ion sources for life science, as well as some of its specific characteristics. To help with the dissemination of the technology, we are also preparing a manuscript detailing the protocol we have used, along with tips for setting up this type of equipment.’
A key part of the research involved evaluating and refining the quality of the plasma-based technique and images, and automating the workflow. Study co-author Dr Elaine Ho, Research Software Engineer in the Franklin’s AI and Informatics theme, said: ‘Our job in the AI and Informatics theme is to build tools that help scientists make use of their data. The tools we built here helped the researchers to evaluate their images and figure out the best protocol going forward. We looked both at how to measure the quality of the images produced and whether we could do anything to improve that quality.’
Their first tool evaluated image resolution to identify the smallest visible biological feature within a sample, while the second measured the ‘curtaining’ – streaks across the image – created by different variations of the technique being tested. A third, machine learning-based tool was designed to remove shadows caused when parts of a sample hold on to their electrical charge. Another tool to align the 3D images based on the physical properties of the microscope was also developed.
Dr Ho added: ‘What’s novel about our work here is that we’ve developed these image processing tools so they can be used by researchers across multiple microscopy techniques without the need to be able to code. We’ve created a software plug-in for our image resolution tool and have packaged all three tools together as an open resource for the scientific community. It’s really important to us that our work has a life beyond this individual study.’
The team has named this toolkit Okapi-EM after a relative of the giraffe whose striped markings only cover part of its body – reminiscent of the removal of streaky artefacts from the cryo-plasma FIB/SEM images.
Franklin researchers collaborated on this study with scientists from the Max Planck Institute of Biochemistry, Thermo Fisher Scientific, Diamond Light Source, and the Mary Lyon Centre at MRC Harwell.
References
Perdigao et al. - Okapi-EM: A napari plugin for processing and analyzing cryogenic serial focused ion beam/scanning electron microscopy images - Biological Imaging (2023)
Related publication
Dumoux et al. - Cryo-plasma FIB/SEM volume imaging of biological specimens - eLife (2023)